Projects: Group B
Group B : Dynamics of multi-protein complexes
Title: Structure and dynamics of the mitochondrial presequence translocase
Principal Investigator: Prof. Dr. Peter Rehling 
Summary: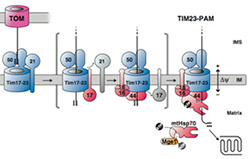 The aim of the project is to structurally and functionally dissect presequence receptor domains and to analyze the dynamics of the translocation machinery in order to understand how protein interactions within the translocase promote the vectorial movement of the precursor protein across the inner and outer mitochondrial membranes.
The aim of the project is to structurally and functionally dissect presequence receptor domains and to analyze the dynamics of the translocation machinery in order to understand how protein interactions within the translocase promote the vectorial movement of the precursor protein across the inner and outer mitochondrial membranes.
Title: Coupled binding and folding in protein assemblies
Principal Investigator: Prof. Dr. Markus Zweckstetter 
Summary:
 Intrinsically disordered proteins (IDPs) and protein regions play important roles in assembly and regulation of dynamic multiprotein complexes and liquid-liquid phase separated states. The dynamic nature of IDPs, however, poses a huge challenge for characterization of their conformations and dynamics. I propose to use NMR spectroscopy to solve the 3D structure of the inner mitochondrial Tim50IMS/Tim23IMS complex and its modulation by presequences and Tim17IMS. In addition, I plan to study the conformations and phase separations of the intrinsically disordered C-terminal region of RNA polymerase II.
Intrinsically disordered proteins (IDPs) and protein regions play important roles in assembly and regulation of dynamic multiprotein complexes and liquid-liquid phase separated states. The dynamic nature of IDPs, however, poses a huge challenge for characterization of their conformations and dynamics. I propose to use NMR spectroscopy to solve the 3D structure of the inner mitochondrial Tim50IMS/Tim23IMS complex and its modulation by presequences and Tim17IMS. In addition, I plan to study the conformations and phase separations of the intrinsically disordered C-terminal region of RNA polymerase II.
Title: Structure and molecular function of the WD-40 repeat containing autophagy proteins Atg18, Atg21 and Hsv2
Principal Investigator: Prof. Dr. Michael Thumm  , Dr. Karin Kühnel
, Dr. Karin Kühnel 
Summary:
Atg18, Atg21 and Hsv2 belong to the PROPPIN family (-propellers that bind PIPs), which differently affect autophagic subtypes. -propeller proteins are platforms for multi-protein complexes. We want to dissect the composition, structure and molecular function of Atg18, Atg21 and Hsv2 complexes and understand their dynamic assembly and disassembly. In the crystal structure of KlHsv2 we uncovered two PI3P-binding sites. Now, we show that Atg21 binds via PI3P to growing autophagosomes, where it arranges PI3P-dependent the Atg8 lipidation machinery in its functional orientation.
Title: Assembly and activation of multimolecular signaling clusters in B lymphocytes
Principal Investigator: Prof. Dr. Jürgen Wienands  , Prof. Dr. Christian Griesinger
, Prof. Dr. Christian Griesinger 
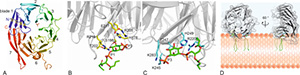
Summary: Antibody-mediated immune responses are initiated upon ligation of the B cell antigen receptor which activates intracellular signal chains by virtue of the adaptor protein SLP65. We discovered that the signaling competence of SLP65 requires a three-step multimerization process resulting in the formation of phase-separated supra-molecular clusters. The assembly of these cytosolic 'signalosomes' is driven by the SH3 domain scaffolding protein CIN85. Our project aims at corrobarating the developing concept of biophysical compart¬mentalization of intracellular signaling elements through phase separation.
Antibody-mediated immune responses are initiated upon ligation of the B cell antigen receptor which activates intracellular signal chains by virtue of the adaptor protein SLP65. We discovered that the signaling competence of SLP65 requires a three-step multimerization process resulting in the formation of phase-separated supra-molecular clusters. The assembly of these cytosolic 'signalosomes' is driven by the SH3 domain scaffolding protein CIN85. Our project aims at corrobarating the developing concept of biophysical compart¬mentalization of intracellular signaling elements through phase separation.
Title: Dynamics of the fungal COP9 signalosome and lid MPN/PCI
Principal Investigator: Prof. Dr. Gerhard Braus 
Summary:
Our focus in the next funding period will be the sequential order of the assembly of the fungal COP9 signalosome (CSN) in vivo from a trimeric and heptameric pre-CSN to a functional CSN. Posttranslational modifications in the assembly of CSN and of the lid of the 19S regulatory particle of the proteasome will be compared. Dynamics of CSN and lid supercomplexes with SemA, the fungal CandA complex of A. nidulans or E3 SCF ubiquitin ligases be addressed.
Title: Structure-function analysis of nuclear export complexes and their interaction with nucleoporins
Principal Investigator: Prof. Dr. Ralph Kehlenbach  , Prof. Dr. Ralf Ficner
, Prof. Dr. Ralf Ficner 
Summary: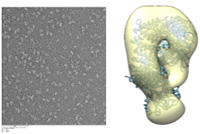 CRM1 is the major nuclear export receptor for proteins and also for a subset of RNAs. In the first part of this project, we plan to analyse the significance of the interaction of CRM1 with an NES-like sequence in the nucleoporin Nup214 for nuclear transport and also for the biogenesis of the nuclear pore complex. In the second part, we want to investigate CRM1-containing RNA-export complexes by means of single-particle cryo-electron microscopy (cryo-EM), X-ray crystallography and mass spectrometry.
CRM1 is the major nuclear export receptor for proteins and also for a subset of RNAs. In the first part of this project, we plan to analyse the significance of the interaction of CRM1 with an NES-like sequence in the nucleoporin Nup214 for nuclear transport and also for the biogenesis of the nuclear pore complex. In the second part, we want to investigate CRM1-containing RNA-export complexes by means of single-particle cryo-electron microscopy (cryo-EM), X-ray crystallography and mass spectrometry.
Title: Computational methods for modeling macromolecular complexes
Principal Investigator: Dr. Michael Habeck 
Summary:
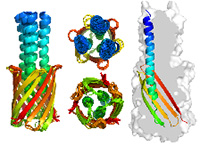 Project B09 aims to develop (1) inference methods to reconstruct models of structural dynamics from cryo-EM data and (2) computational methods for coarse-grained modeling of macromolecular complexes. To realize aim (1), we will implement various probabilistic models to describe structural dynamics and develop sampling algorithms to infer motion models from 3D and 2D data. To realize aim (2), we will support coarse-grained representations of biomolecular assemblies in our ISD software and develop sampling algorithms for rigid and flexible fitting with hybrid structural data.
Project B09 aims to develop (1) inference methods to reconstruct models of structural dynamics from cryo-EM data and (2) computational methods for coarse-grained modeling of macromolecular complexes. To realize aim (1), we will implement various probabilistic models to describe structural dynamics and develop sampling algorithms to infer motion models from 3D and 2D data. To realize aim (2), we will support coarse-grained representations of biomolecular assemblies in our ISD software and develop sampling algorithms for rigid and flexible fitting with hybrid structural data.
Title: The role of phosphorylations in intermediate filament assembly and disassembly
Principal Investigator: Prof. Dr. Sarah Köster 
Summary:
Intermediate filaments (IFs) form very stable macromolecular structures in the cytoskeleton. In the cell, the assembly and especially the disassembly reaction of these extended protein filaments is regulated by posttranslational phophorylations. We propose to study IF disassembly upon addition of kinases and in a phosphomimicri approach by targeted mutagenesis. We will focus in vimentin, which is the most abundant IF protein, and employ small angle x-ray scattering (SAXS) as the main method. Our aim is to shed light on the mechanisms and time scales involved in IF disassembly.
Title: Structure and dynamics of the NPC permeability barrier
Principal Investigator: Dr. Loren Andreas  , Prof. Dirk Görlich
, Prof. Dirk Görlich 
Summary:
Nuclear pore complexes (NPCs) conduct nucleocytoplasmic transport through a selective permeability barrier formed by cohesive and intrinsically disordered FG repeat domains. Through a combined analysis by crystallography, biophysical characterization, and NMR spectroscopy, we will investigate the structure and dynamics of barrier-forming units as well as the detailed mechanisms of sieving and privileged nuclear transport receptor passage. Thereby we wish to understand the biophysical determinants of effective transport.
Contact
Spokesperson
Prof. Dr. Ralf Ficner 
Telfon: +49 (0)551-39-28618
Fax: +49 (0)551-39-28629
SFB860 Administration
Contact email: 
Susanne van Beckum 
Tel.: +49 (0)551-39-28611
Fax: +49 (0)551-39-28629
SFB860
Georg-August-Universität Göttingen
Abtl. Molekulare Strukturbiologie
Justus-von-Liebig-Weg 11
37077 Göttingen


