Projects: Group A
Group A : Dynamics of ribonucleoprotein complexes
Title: Crystallographic studies on molecular motors of the spliceosome
Principal Investigator: Prof. Dr. Ralf Ficner 
Summary:
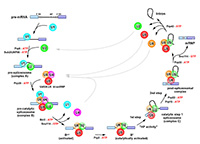 Eight different DExD/H-box ATPases promote the structural and compositional changes required for the assembly, activation and disassembly of the spliceosome. The activities of the DEAH-box proteins Prp2 and Prp43 in the spliceosome are specifically triggered by direct interaction with the intrinsically disordered G-patch domains of the proteins Spp2 and Ntr1, respectively, while Prp22 exhibits a high intrinsic activity. We aim to unravel the structural basis for the stimulation by G-patch domains, as well as the molecular mechanism of RNA unwinding by Prp43 and Prp22.
Eight different DExD/H-box ATPases promote the structural and compositional changes required for the assembly, activation and disassembly of the spliceosome. The activities of the DEAH-box proteins Prp2 and Prp43 in the spliceosome are specifically triggered by direct interaction with the intrinsically disordered G-patch domains of the proteins Spp2 and Ntr1, respectively, while Prp22 exhibits a high intrinsic activity. We aim to unravel the structural basis for the stimulation by G-patch domains, as well as the molecular mechanism of RNA unwinding by Prp43 and Prp22.
Title: Ribosome dynamics in translation
Principal Investigator: Prof. Dr. Marina Rodnina 
Summary:
During protein synthesis, ribosome, tRNAs, and protein factors move in a precisely orchestrated way to translate mRNA into a functional polypeptide. We will use biophysical techniques (ensemble and single molecule fluorescence and FRET) in combination with structural analysis to investigate the conformational and compositional dynamics of the ribosome–tRNA–translation factor complexes at each step of translation. We will analyse and how ribosome dynamics change in response to translation pausing and recoding events. This work will reveal how spontaneous Brownian motions of molecules are translated into biological function.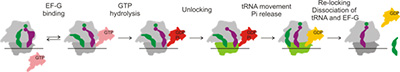
Title: High-resolution structure determination of macromolecular complexes by cryo-EM
Principal Investigator: Prof. Dr. Holger Stark 
Summary:
The planned work program includes the high-resolution 3D structure determination of several macromolecular complexes, which are current projects in the SFB860. The focus for the next funding period will be on structure determination of further intermediate states of the dynamic spliceosome. The quantitative analysis of the dynamics of the spliceosome is of particular interest, for which we have planned methodological work to improve computer-aided image processing procedures. Another project is the structural analysis of the human 48S Translation-Initiations complex.
Title: Single-molecule fluorescence spectroscopy and imaging of the structure and dynamics of macromolecular assemblies
Principal Investigator: Prof. Dr. Jörg Enderlein 
Summary: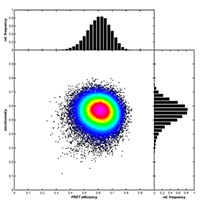 The core goal of the project is the development of a fluorescence-optical method for three-dimensional localization and co-localization of single fluorescent molecules with ~1 nm isotropic resolution. For this purpose, we will combine two techniques: single-molecule localization based super-resolution microscopy (STORM, PALM, PAINT, MinFlux) with metal Induced Energy Transfer (MIET) imaging. We will use this method to image structural details of fluorescently labelled macromolecular complexes with nanometer resolution and high specificity, but also to resolve temporal dynamics of structural changes with a temporal resolution down to a few nanoseconds.
The core goal of the project is the development of a fluorescence-optical method for three-dimensional localization and co-localization of single fluorescent molecules with ~1 nm isotropic resolution. For this purpose, we will combine two techniques: single-molecule localization based super-resolution microscopy (STORM, PALM, PAINT, MinFlux) with metal Induced Energy Transfer (MIET) imaging. We will use this method to image structural details of fluorescently labelled macromolecular complexes with nanometer resolution and high specificity, but also to resolve temporal dynamics of structural changes with a temporal resolution down to a few nanoseconds.
Title: Protein cross-linking of macromolecular complexes
Principal Investigator: Dr. Henning Urlaub 
Summary: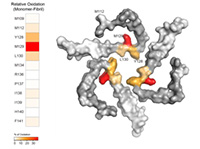 We will perform protein cross-linking combined with mass spectrometry (MS) of resting and stimulated B cells. The cross-linked interactome of key signaling molecules will be related to the phosphorylation states of the B cells upon B cell receptor (BCR) stimulation and information on how phosphorylation influences molecular interactions will be obtained. Further, we will establish protein-DNA cross-linking combined with MS of in vitro reconstituted protein-DNA complexes and in in vivo and ex vivo systems using UV-light and chemical cross-linker. Structural function relationships in DNA–protein systems like chromatin will be obtained.
We will perform protein cross-linking combined with mass spectrometry (MS) of resting and stimulated B cells. The cross-linked interactome of key signaling molecules will be related to the phosphorylation states of the B cells upon B cell receptor (BCR) stimulation and information on how phosphorylation influences molecular interactions will be obtained. Further, we will establish protein-DNA cross-linking combined with MS of in vitro reconstituted protein-DNA complexes and in in vivo and ex vivo systems using UV-light and chemical cross-linker. Structural function relationships in DNA–protein systems like chromatin will be obtained.
Title: Molecular mechanisms and complex formation in the cytoplasmic mRNA quality control
Principal Investigator: Prof. Dr. Heike Krebber 
 Summary:
Summary:
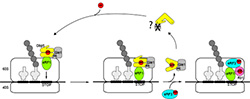
In this project we will investigate the mechanisms of the cytoplasmic mRNA quality control systems. We have identified the DEAD-box RNA helicase Dbp5 and the nuclear guard-proteins Gbp2 and Hrb1 to be novel players in these cytoplasmic quality control pathways. This project aims to characterize their function in these processes by analysis of their in vivo complex formations in different mutants of the model organism Saccharomyces cerevisiae.
Title: RNA elements and protein factors required for pre-ribosome remodelling during 60S biogenesis in yeast
Principal Investigator: Prof. Dr. Markus T. Bohnsack 
Summary:
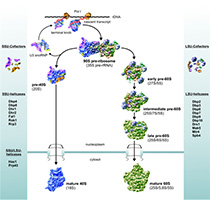
Ribosome biogenesis in eukaryotes is a major cellular pathway that requires multiple cofactors, among them DEAD/H-box RNA helicases. We have identified binding sites of multiple large subunit RNA helicases on yeast ribosomal RNAs and analyse their molecular functions in the pathway. Our data also reveal interactions between several helicases and non-ribosomal RNAs, suggesting that these proteins perform additional functions in other pathways. Understanding the molecular functions and regulation of RNA helicases as well as their interplay will shed light on key steps in RNA metabolism and the cross-regulation of different cellular processes.
Title: lntegrated structural biology of the Mediator complex
Principal Investigator: Prof. Dr. Patrick Cramer 
Summary:
The aim of this project is to solve the structure of the Mediator complex, the central coactivator for RNA polymerase (Pol) II transcription. During the current funding period, we solved the structure of the 15-subunit core Mediator (Nozawa et al., Nature 2017). During the next funding period, we wish to extend the structure to the 21-subunit Mediator by a combination of co-expression of Mediator subunits, cryo-electron microscopy and X-ray crystallography.
Title: Translation termination in human mitochondria: Why do we need four release factors?
Principal Investigator: Dr. Ricarda Richter-Dennerlein 
Summary:
In human mitochondria there are four predicted class I release factors (RFs): mtRF1a, mtRF1, ICT1 and C12ORF65. However, a codon-dependent translation termination activity has been only demonstrated for mtRF1a so far. It has been suggested that mtRF1a is sufficient to terminate all mitochondrial transcripts. The role of the other RFs in mitochondrial translation termination remains elusive. Here, we will investigate the function of mtRF1, which shows significant differences in the decoding motifs compared to other RFs.
Title: Scanning dynamics of the mammalian ribosome during translation initiation
Principal Investigator: Dr. Sarah Adio 
Summary:
Translation of most eukaryotic mRNAs starts by a scanning mechanism. The small ribosomal subunit is recruited to the mRNA 5’ end and then migrates in 3’ direction towards the AUG start codon. Cells have developed multiple mechanisms to regulate the scanning process and thus shape their proteome. Misregulation can lead to diseases such as cancer and viral infections. We will study scanning using single molecule Förster Resonance Energy Transfer (smFRET) and Total Internal Reflection of Fluorescence (TIRF) microscopy. Our work will establish the mechanisms of function and regulation of gene expression during translation in eukaryotes.
Title: Molecular dynamics simulations of protein-nucleotide interaction
Principal Investigator: Prof. Dr. Jochen Hub 
Summary:
We will use molecular dynamics (MD) simulations to study conformational transitions of RNA polymerase II (Pol II) and of several members of the RNA helicase family. Specifically, we aim to derive energetically and sterically feasible pathways for DNA opening and loading into the catalytic cleft of Pol II. By means of SAXS-guided MD simulations and free energy calculations, we aim to reveal the mechanisms involved in RNA loading and RNA sliding by RNA helicases.
Contact
Spokesperson
Prof. Dr. Ralf Ficner 
Tel.: +49 (0)551-39-28618
Fax: +49 (0)551-39-28629
SFB860 Administration
Contact email: 
Susanne van Beckum 
Tel.: +49 (0)551-39-28611
Fax: +49 (0)551-39-28629
SFB860
Georg-August-Universität Göttingen
Abtl. Molekulare Strukturbiologie
Justus-von-Liebig-Weg 11
37077 Göttingen


